Georg Steinert
marine ecology and systematics
The Mytilus complex in the Wadden Sea of Germay (2008 - 2010)
Project participants: Georg Steinert, Olaf R.P. Bininda-Emonds, Funding: EWE-FoundationThe publication "Species status and population structure of mussels (Mollusca: Bivalvia: Mytilus spp.) in the Wadden Sea of Lower Saxony (Germany)" is available on SpringerLink


Fig. 1 & Fig. 2. M. edulis specimen from an intertidal blue mussel bed located in the Wadden Sea of Germany
Three species of the blue mussel, Mytilus, occur in the North Sea region: M. edulis (Fig. 1 & 2), M. galloprovincialis and M. trossulus (Gosling 1992). Hybridization is likely to occur in areas where the their distributions overlap. The blue mussel (M. edulis) is supposed to be the indigenous species in the Lower Saxonian Wadden Sea. Recent studies examined the taxonomic status and genetic composition of Mytilus populations in the Netherlands, Great Britain and the Baltic Sea in general. However, to my knowledge comparable genetic studies in the German Wadden Sea of this genus were never carried out.
A total of 504 mussels from 15 sample areas were assayed for the Nuclear DNA gentic marker Me 15/16 (Fig. 3). I analyzed the mussels genetic structure using phylo- and populationgenetic methods. Therefore two fragments of maternal transmitted mitochondrial DNA, comprising parts of cytochrome c oxidase subunit one and the first variable domain of the control region were sequenced from >120 individuals (Fig. 4). The mussels were identified as 99% M. edulis and 1% M. edulis / galloprovincialis hybrids composed of two mixed genotypes (Fig. 3). Overall, genetic composition results showed a heterogeneous population structure due to unrestricted gene flow within the sampling area.
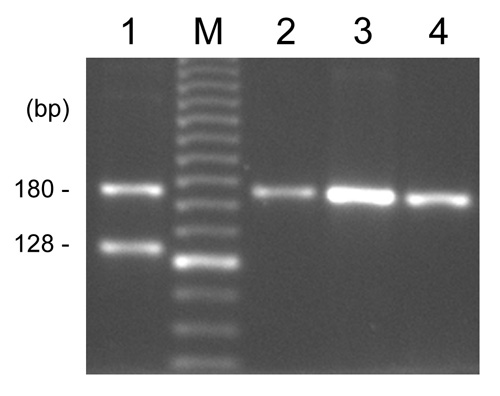
Fig. 3. Example of the Me1516 PCR results. The size of the amplified fragment is 180 bp in M. edulis and 128 bp in M. galloprovincialis. Two fragments are presumed to be hybrids between the two species (e.g. lane 1).

Fig. 4. VD1 and CO1 primer sites and fragment design. Different fragment length of the maternal (F-mtDNA) mitochondrial control region (D-Loop). Also shown primer sites VD1F, VD1R, MytF and MytR (Wares and Cunnigham 2001) used in the present study. 16S 16S ribosomal RNA; VD1 Variable Domain 1; CD Constant Domain; VD2 Variable Domain 2; Y Tyrosin; COI Cytochrome-c-Oxidase Subunit 1
This is the first record of the presence of hybrid Mytilus mussels in the North Saxonian Wadden Sea. Additionally, this is the first genetic demonstration of a M. edulis panmictic population structure of the North Saxonian intertidal. The observed heterogenous population can be attributed to extensive larval dispersal linked to the prevailing hydrodynamic conditions in the intertidal back barrier basins of the Wadden Sea.
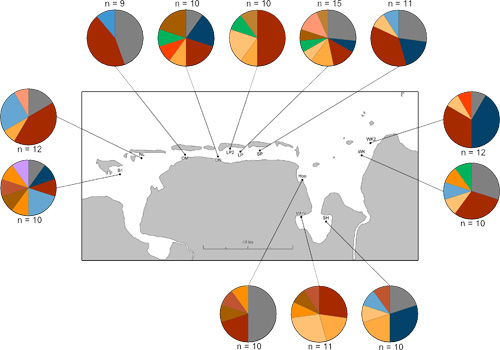
Fig. 5. Diversity and distribution of the Cytochrome-c-Oxidase Subunit 1 (COI) fragment haplotypes. The grey color represents the unique n = 1 haplotypes within the mitochondrial sequence data set. The other colors are related to the n > 2 haplotypes within the data set. The colors match with the colouring of the COI haplotype network (Fig. 6).
Applied statistical methods and software-tools
Phylogenetic analysis
- Haplotype-Networks with TCS 2.1 (Fig. 6)
- Maximum Likelihood trees with RAxML 7.0.4 (Fig. 7)
- Maximum Parsimony trees with PAUP* 4.0b10
Population genetics
- Analysis of Molecular Variance (AMOVA) with Arlequin 3.11
- Pairwise difference with Arlequin 3.11
- Clusteranalyses with PAST 1.97 (Fig. 8)
Recombionation detection with RDP3 (Recombination Detection Programm)

Fig. 6. The sizes of the circles and the rectangle are proportional to the number of individuals sharing that haplotype (n = 1 not indicated). The haplotype in the rectangle has the biggest outgroup weight. Each node corresponds to one mutation step. The colors refer to the COI geographical distribution map (Fig. 5).

Fig. 7. Phylogenetic tree of COI maternal mitochondrial haplotypes of the Wadden Sea M. edulis based on maximum likelihood estimates constructed using RAxML. The icons highlight the position of the GenBank sequences within the tree. Genealogy based on the general time reversible model of evolution. 1000 replicates.

Fig. 8. Clusteranalysis with PAST
References
- Gosling E. (1992). Genetics of Mytilus. In: 'The mussel Mytilus: ecology, physiology genetics and culture'. E. Gosling (ed), Elsevier Amsterdam, 309-382.
- Inoue, K., J.H. Waite, M. Matsuoka, S. Odo, S. Harayama (1995). Interspecific variations in adhesive protein gene squencies of Mytilus edulis, M. galloprovincialis and M. trossulus. Biological Bulletin 189:370-375.
- Kijewski, T., J. W. M. Wijsman, et al. (2009). Genetic composition of cultured and wild mussels Mytilus from the Netherlands and transfers from Ireland and Great Britain. Aquaculture 287(3-4): 292-296.
- Luttikhuizen, P.C., A. Koolhaas, et al. (2002). Mytilus galloprovincialis-type foot-protein-1 alleles occur at low frequency among mussels in the Dutch Wadden Sea. Journal of Sea Research 48(3): 241-245.
- Wares, J. P., & Cunningham, C. W. (2001). Phylogeography and historical ecology of the North Atlantic intertidal. Evolution, 55(12), 2455-2469.